Visualization with hierarchical clustering and t-SNE
Contents
Visualization with hierarchical clustering and t-SNE#
In this chapter, you’ll learn about two unsupervised learning techniques for data visualization, hierarchical clustering and t-SNE. Hierarchical clustering merges the data samples into ever-coarser clusters, yielding a tree visualization of the resulting cluster hierarchy. t-SNE maps the data samples into 2d space so that the proximity of the samples to one another can be visualized.
Visualizing hierarchies#
How many merges?#
If there are 5 data samples, how many merge operations will occur in a hierarchical clustering? (To help answer this question, think back to the video, in which Ben walked through an example of hierarchical clustering using 6 countries.)
4 merges.
3 merges.
This can’t be known in advance.
Hierarchical clustering of the grain data#
In the video, you learned that the SciPy linkage() function
performs hierarchical clustering on an array of samples. Use the
linkage() function to obtain a hierarchical clustering of
the grain samples, and use dendrogram() to visualize the
result. A sample of the grain measurements is provided in the array
samples, while the variety of each grain sample is given by
the list varieties.
linkage and dendrogram from
scipy.cluster.hierarchy.
matplotlib.pyplot as plt.
samples using the
linkage() function with the method=‘complete’
keyword argument. Assign the result to mergings.
dendrogram() function on
mergings. Specify the keyword arguments
labels=varieties, leaf_rotation=90, and
leaf_font_size=6.
# edited/added
samples = np.array(grains.sample(42))[:,:7]
varieties = list(np.array(grains.sample(42))[:,8])
# Perform the necessary imports
from scipy.cluster.hierarchy import linkage, dendrogram
import matplotlib.pyplot as plt
# Calculate the linkage: mergings
mergings = linkage(samples, method='complete')
# Plot the dendrogram, using varieties as labels
dendrogram(mergings,
labels=varieties,
leaf_rotation=90,
leaf_font_size=6,
)
## {'icoord': [[25.0, 25.0, 35.0, 35.0], [15.0, 15.0, 30.0, 30.0], [55.0, 55.0, 65.0, 65.0], [45.0, 45.0, 60.0, 60.0], [22.5, 22.5, 52.5, 52.5], [5.0, 5.0, 37.5, 37.5], [85.0, 85.0, 95.0, 95.0], [105.0, 105.0, 115.0, 115.0], [125.0, 125.0, 135.0, 135.0], [110.0, 110.0, 130.0, 130.0], [145.0, 145.0, 155.0, 155.0], [165.0, 165.0, 175.0, 175.0], [150.0, 150.0, 170.0, 170.0], [120.0, 120.0, 160.0, 160.0], [90.0, 90.0, 140.0, 140.0], [75.0, 75.0, 115.0, 115.0], [185.0, 185.0, 195.0, 195.0], [205.0, 205.0, 215.0, 215.0], [225.0, 225.0, 235.0, 235.0], [255.0, 255.0, 265.0, 265.0], [245.0, 245.0, 260.0, 260.0], [230.0, 230.0, 252.5, 252.5], [210.0, 210.0, 241.25, 241.25], [190.0, 190.0, 225.625, 225.625], [275.0, 275.0, 285.0, 285.0], [305.0, 305.0, 315.0, 315.0], [295.0, 295.0, 310.0, 310.0], [280.0, 280.0, 302.5, 302.5], [207.8125, 207.8125, 291.25, 291.25], [345.0, 345.0, 355.0, 355.0], [335.0, 335.0, 350.0, 350.0], [325.0, 325.0, 342.5, 342.5], [375.0, 375.0, 385.0, 385.0], [405.0, 405.0, 415.0, 415.0], [395.0, 395.0, 410.0, 410.0], [380.0, 380.0, 402.5, 402.5], [365.0, 365.0, 391.25, 391.25], [333.75, 333.75, 378.125, 378.125], [249.53125, 249.53125, 355.9375, 355.9375], [95.0, 95.0, 302.734375, 302.734375], [21.25, 21.25, 198.8671875, 198.8671875]], 'dcoord': [[0.0, 0.35892177420713867, 0.35892177420713867, 0.0], [0.0, 0.6444799531405162, 0.6444799531405162, 0.35892177420713867], [0.0, 0.6327327160816018, 0.6327327160816018, 0.0], [0.0, 1.57472866234155, 1.57472866234155, 0.6327327160816018], [0.6444799531405162, 2.1906390300549288, 2.1906390300549288, 1.57472866234155], [0.0, 3.8753454671809577, 3.8753454671809577, 2.1906390300549288], [0.0, 0.7207984184222385, 0.7207984184222385, 0.0], [0.0, 0.3449266008877837, 0.3449266008877837, 0.0], [0.0, 0.5373874300725677, 0.5373874300725677, 0.0], [0.3449266008877837, 1.0132117251591595, 1.0132117251591595, 0.5373874300725677], [0.0, 0.4019535296523724, 0.4019535296523724, 0.0], [0.0, 0.74000216891574, 0.74000216891574, 0.0], [0.4019535296523724, 1.1923667892054022, 1.1923667892054022, 0.74000216891574], [1.0132117251591595, 1.7057229229860276, 1.7057229229860276, 1.1923667892054022], [0.7207984184222385, 2.76135637142329, 2.76135637142329, 1.7057229229860276], [0.0, 4.8292916664869185, 4.8292916664869185, 2.76135637142329], [0.0, 0.8213969137998998, 0.8213969137998998, 0.0], [0.0, 0.7680247456950849, 0.7680247456950849, 0.0], [0.0, 0.39210953826705036, 0.39210953826705036, 0.0], [0.0, 0.44020218082149526, 0.44020218082149526, 0.0], [0.0, 0.6878648122996261, 0.6878648122996261, 0.44020218082149526], [0.39210953826705036, 0.8667876095099653, 0.8667876095099653, 0.6878648122996261], [0.7680247456950849, 1.6315417279371072, 1.6315417279371072, 0.8667876095099653], [0.8213969137998998, 2.0021865947009037, 2.0021865947009037, 1.6315417279371072], [0.0, 0.7600501365041651, 0.7600501365041651, 0.0], [0.0, 0.4355730593138186, 0.4355730593138186, 0.0], [0.0, 0.8899149172814218, 0.8899149172814218, 0.4355730593138186], [0.7600501365041651, 2.2825246898993226, 2.2825246898993226, 0.8899149172814218], [2.0021865947009037, 3.108696229933058, 3.108696229933058, 2.2825246898993226], [0.0, 0.28514601522728705, 0.28514601522728705, 0.0], [0.0, 1.0559243533511296, 1.0559243533511296, 0.28514601522728705], [0.0, 2.196101828240212, 2.196101828240212, 1.0559243533511296], [0.0, 0.5513404120867617, 0.5513404120867617, 0.0], [0.0, 0.5540343761897811, 0.5540343761897811, 0.0], [0.0, 0.8413507235392381, 0.8413507235392381, 0.5540343761897811], [0.5513404120867617, 1.6519585981494824, 1.6519585981494824, 0.8413507235392381], [0.0, 2.6224088640027143, 2.6224088640027143, 1.6519585981494824], [2.196101828240212, 4.713556156449182, 4.713556156449182, 2.6224088640027143], [3.108696229933058, 5.923029677453931, 5.923029677453931, 4.713556156449182], [4.8292916664869185, 8.159839861173747, 8.159839861173747, 5.923029677453931], [3.8753454671809577, 10.666543593873321, 10.666543593873321, 8.159839861173747]], 'ivl': ['Kama wheat', 'Kama wheat', 'Rosa wheat', 'Kama wheat', 'Rosa wheat', 'Rosa wheat', 'Rosa wheat', 'Kama wheat', 'Canadian wheat', 'Canadian wheat', 'Canadian wheat', 'Rosa wheat', 'Canadian wheat', 'Rosa wheat', 'Rosa wheat', 'Kama wheat', 'Canadian wheat', 'Rosa wheat', 'Kama wheat', 'Canadian wheat', 'Kama wheat', 'Canadian wheat', 'Rosa wheat', 'Canadian wheat', 'Canadian wheat', 'Rosa wheat', 'Canadian wheat', 'Rosa wheat', 'Rosa wheat', 'Rosa wheat', 'Canadian wheat', 'Kama wheat', 'Kama wheat', 'Canadian wheat', 'Kama wheat', 'Rosa wheat', 'Rosa wheat', 'Canadian wheat', 'Rosa wheat', 'Rosa wheat', 'Kama wheat', 'Canadian wheat'], 'leaves': [32, 8, 6, 16, 39, 1, 34, 10, 9, 29, 14, 26, 5, 18, 3, 22, 7, 11, 13, 21, 15, 31, 28, 38, 20, 19, 30, 0, 33, 4, 2, 41, 40, 17, 27, 37, 36, 24, 35, 25, 12, 23], 'color_list': ['C1', 'C1', 'C1', 'C1', 'C1', 'C1', 'C2', 'C2', 'C2', 'C2', 'C2', 'C2', 'C2', 'C2', 'C2', 'C2', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C3', 'C0', 'C0']}
plt.show()
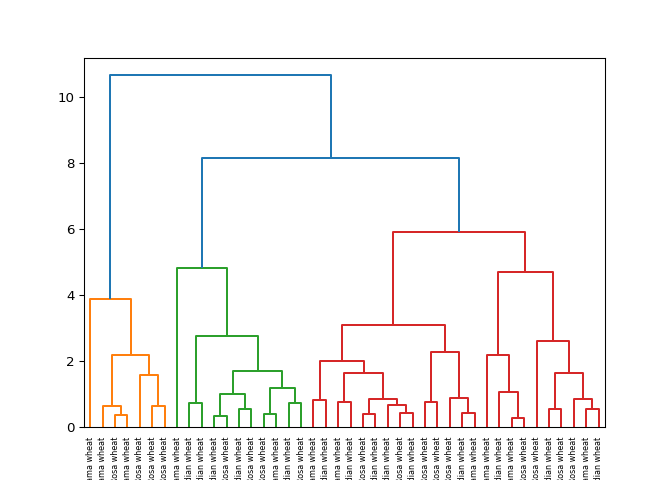
Superb! Dendrograms are a great way to illustrate the arrangement of the clusters produced by hierarchical clustering.
Hierarchies of stocks#
In chapter 1, you used k-means clustering to cluster companies according
to their stock price movements. Now, you’ll perform hierarchical
clustering of the companies. You are given a NumPy array of price
movements movements, where the rows correspond to
companies, and a list of the company names companies. SciPy
hierarchical clustering doesn’t fit into a sklearn pipeline, so you’ll
need to use the normalize() function from
sklearn.preprocessing instead of Normalizer.
linkage and dendrogram have already been
imported from scipy.cluster.hierarchy, and PyPlot has been
imported as plt.
normalize from sklearn.preprocessing.
normalize() function on movements.
linkage() function to
normalized_movements, using ‘complete’
linkage, to calculate the hierarchical clustering. Assign the result to
mergings.
companies of company names as the labels. In
addition, specify the leaf_rotation=90, and
leaf_font_size=6 keyword arguments as you did in the
previous exercise.
# Import normalize
from sklearn.preprocessing import normalize
# Normalize the movements: normalized_movements
normalized_movements = normalize(movements)
# Calculate the linkage: mergings
mergings = linkage(normalized_movements, method='complete')
# Plot the dendrogram
dendrogram(
mergings,
labels=companies,
leaf_rotation=90,
leaf_font_size=6
)
## {'icoord': [[25.0, 25.0, 35.0, 35.0], [15.0, 15.0, 30.0, 30.0], [45.0, 45.0, 55.0, 55.0], [75.0, 75.0, 85.0, 85.0], [65.0, 65.0, 80.0, 80.0], [50.0, 50.0, 72.5, 72.5], [22.5, 22.5, 61.25, 61.25], [5.0, 5.0, 41.875, 41.875], [105.0, 105.0, 115.0, 115.0], [95.0, 95.0, 110.0, 110.0], [175.0, 175.0, 185.0, 185.0], [165.0, 165.0, 180.0, 180.0], [155.0, 155.0, 172.5, 172.5], [145.0, 145.0, 163.75, 163.75], [205.0, 205.0, 215.0, 215.0], [235.0, 235.0, 245.0, 245.0], [225.0, 225.0, 240.0, 240.0], [210.0, 210.0, 232.5, 232.5], [195.0, 195.0, 221.25, 221.25], [154.375, 154.375, 208.125, 208.125], [295.0, 295.0, 305.0, 305.0], [285.0, 285.0, 300.0, 300.0], [275.0, 275.0, 292.5, 292.5], [265.0, 265.0, 283.75, 283.75], [345.0, 345.0, 355.0, 355.0], [335.0, 335.0, 350.0, 350.0], [325.0, 325.0, 342.5, 342.5], [315.0, 315.0, 333.75, 333.75], [274.375, 274.375, 324.375, 324.375], [255.0, 255.0, 299.375, 299.375], [181.25, 181.25, 277.1875, 277.1875], [135.0, 135.0, 229.21875, 229.21875], [125.0, 125.0, 182.109375, 182.109375], [102.5, 102.5, 153.5546875, 153.5546875], [23.4375, 23.4375, 128.02734375, 128.02734375], [365.0, 365.0, 375.0, 375.0], [395.0, 395.0, 405.0, 405.0], [425.0, 425.0, 435.0, 435.0], [415.0, 415.0, 430.0, 430.0], [400.0, 400.0, 422.5, 422.5], [385.0, 385.0, 411.25, 411.25], [370.0, 370.0, 398.125, 398.125], [465.0, 465.0, 475.0, 475.0], [455.0, 455.0, 470.0, 470.0], [445.0, 445.0, 462.5, 462.5], [485.0, 485.0, 495.0, 495.0], [535.0, 535.0, 545.0, 545.0], [525.0, 525.0, 540.0, 540.0], [515.0, 515.0, 532.5, 532.5], [505.0, 505.0, 523.75, 523.75], [565.0, 565.0, 575.0, 575.0], [555.0, 555.0, 570.0, 570.0], [514.375, 514.375, 562.5, 562.5], [490.0, 490.0, 538.4375, 538.4375], [585.0, 585.0, 595.0, 595.0], [514.21875, 514.21875, 590.0, 590.0], [453.75, 453.75, 552.109375, 552.109375], [384.0625, 384.0625, 502.9296875, 502.9296875], [75.732421875, 75.732421875, 443.49609375, 443.49609375]], 'dcoord': [[0.0, 0.8766150964619032, 0.8766150964619032, 0.0], [0.0, 1.0052496265150068, 1.0052496265150068, 0.8766150964619032], [0.0, 1.0405854841254667, 1.0405854841254667, 0.0], [0.0, 0.9756642737984944, 0.9756642737984944, 0.0], [0.0, 1.0443532462900014, 1.0443532462900014, 0.9756642737984944], [1.0405854841254667, 1.1087706867538036, 1.1087706867538036, 1.0443532462900014], [1.0052496265150068, 1.1539599583553475, 1.1539599583553475, 1.1087706867538036], [0.0, 1.1953921911777048, 1.1953921911777048, 1.1539599583553475], [0.0, 1.0675113301764332, 1.0675113301764332, 0.0], [0.0, 1.162102133265849, 1.162102133265849, 1.0675113301764332], [0.0, 0.6937115853632807, 0.6937115853632807, 0.0], [0.0, 0.7897669950447856, 0.7897669950447856, 0.6937115853632807], [0.0, 0.9060804298422901, 0.9060804298422901, 0.7897669950447856], [0.0, 0.9978691096150067, 0.9978691096150067, 0.9060804298422901], [0.0, 0.850608403749446, 0.850608403749446, 0.0], [0.0, 0.7506255354591677, 0.7506255354591677, 0.0], [0.0, 0.8894957237528659, 0.8894957237528659, 0.7506255354591677], [0.850608403749446, 0.9615815807943434, 0.9615815807943434, 0.8894957237528659], [0.0, 1.0334597062675297, 1.0334597062675297, 0.9615815807943434], [0.9978691096150067, 1.05365593648412, 1.05365593648412, 1.0334597062675297], [0.0, 0.8528392636699416, 0.8528392636699416, 0.0], [0.0, 0.8730000088242031, 0.8730000088242031, 0.8528392636699416], [0.0, 0.9595547992508179, 0.9595547992508179, 0.8730000088242031], [0.0, 0.9933139902556245, 0.9933139902556245, 0.9595547992508179], [0.0, 0.6735509755390415, 0.6735509755390415, 0.0], [0.0, 0.7984090019882444, 0.7984090019882444, 0.6735509755390415], [0.0, 0.8794722330700889, 0.8794722330700889, 0.7984090019882444], [0.0, 1.0410874279228417, 1.0410874279228417, 0.8794722330700889], [0.9933139902556245, 1.0581735997075004, 1.0581735997075004, 1.0410874279228417], [0.0, 1.115596724887607, 1.115596724887607, 1.0581735997075004], [1.05365593648412, 1.1659596038210138, 1.1659596038210138, 1.115596724887607], [0.0, 1.1700635570314206, 1.1700635570314206, 1.1659596038210138], [0.0, 1.1909309430336361, 1.1909309430336361, 1.1700635570314206], [1.162102133265849, 1.236510636000957, 1.236510636000957, 1.1909309430336361], [1.1953921911777048, 1.2467384602108724, 1.2467384602108724, 1.236510636000957], [0.0, 1.1224902285374243, 1.1224902285374243, 0.0], [0.0, 0.9516416111189119, 0.9516416111189119, 0.0], [0.0, 0.9263737672939854, 0.9263737672939854, 0.0], [0.0, 0.9774020092313027, 0.9774020092313027, 0.9263737672939854], [0.9516416111189119, 1.0825651898927742, 1.0825651898927742, 0.9774020092313027], [0.0, 1.1436387132254562, 1.1436387132254562, 1.0825651898927742], [1.1224902285374243, 1.1617351684882877, 1.1617351684882877, 1.1436387132254562], [0.0, 0.804091365214309, 0.804091365214309, 0.0], [0.0, 0.9529466260111008, 0.9529466260111008, 0.804091365214309], [0.0, 1.1381811890921898, 1.1381811890921898, 0.9529466260111008], [0.0, 1.0014106416309905, 1.0014106416309905, 0.0], [0.0, 0.58499795491215, 0.58499795491215, 0.0], [0.0, 0.78536856335178, 0.78536856335178, 0.58499795491215], [0.0, 0.8427599656210099, 0.8427599656210099, 0.78536856335178], [0.0, 0.88159190249364, 0.88159190249364, 0.8427599656210099], [0.0, 0.8547176535466976, 0.8547176535466976, 0.0], [0.0, 0.917129270573618, 0.917129270573618, 0.8547176535466976], [0.88159190249364, 1.0113079675726626, 1.0113079675726626, 0.917129270573618], [1.0014106416309905, 1.0903568915580981, 1.0903568915580981, 1.0113079675726626], [0.0, 1.1116148749505466, 1.1116148749505466, 0.0], [1.0903568915580981, 1.1665446301937066, 1.1665446301937066, 1.1116148749505466], [1.1381811890921898, 1.203388148166268, 1.203388148166268, 1.1665446301937066], [1.1617351684882877, 1.2625272906988811, 1.2625272906988811, 1.203388148166268], [1.2467384602108724, 1.3161900463070635, 1.3161900463070635, 1.2625272906988811]], 'ivl': ['Apple', 'Taiwan Semiconductor Manufacturing', 'Intel', 'Texas instruments', 'Dell', 'HP', 'Symantec', 'Cisco', 'Microsoft', 'Yahoo', 'Amazon', 'Google/Alphabet', 'AIG', 'Valero Energy', 'American express', 'Goldman Sachs', 'Wells Fargo', 'Bank of America', 'JPMorgan Chase', 'Ford', 'Canon', 'Sony', 'Mitsubishi', 'Honda', 'Toyota', 'Navistar', 'IBM', 'General Electrics', '3M', 'Caterpillar', 'DuPont de Nemours', 'Xerox', 'Schlumberger', 'ConocoPhillips', 'Chevron', 'Exxon', 'Home Depot', 'Wal-Mart', 'Philip Morris', 'Coca Cola', 'Pepsi', 'Kimberly-Clark', 'Colgate-Palmolive', 'Procter Gamble', 'Walgreen', 'Boeing', 'Lookheed Martin', 'Northrop Grumman', 'Johnson & Johnson', 'Pfizer', 'SAP', 'Sanofi-Aventis', 'Unilever', 'Royal Dutch Shell', 'Total', 'British American Tobacco', 'GlaxoSmithKline', 'Novartis', 'MasterCard', 'McDonalds'], 'leaves': [0, 50, 24, 51, 14, 22, 47, 11, 33, 59, 2, 17, 1, 53, 3, 18, 55, 5, 26, 15, 7, 45, 34, 21, 48, 35, 23, 16, 32, 8, 13, 58, 44, 10, 12, 57, 20, 56, 41, 28, 38, 27, 9, 40, 54, 4, 29, 36, 25, 39, 43, 46, 52, 42, 49, 6, 19, 37, 30, 31], 'color_list': ['C1', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C2', 'C2', 'C2', 'C0', 'C3', 'C4', 'C4', 'C0', 'C0', 'C0', 'C5', 'C5', 'C0', 'C0', 'C6', 'C6', 'C6', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C7', 'C0', 'C0', 'C0', 'C8', 'C8', 'C8', 'C8', 'C9', 'C9', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0']}
plt.show()
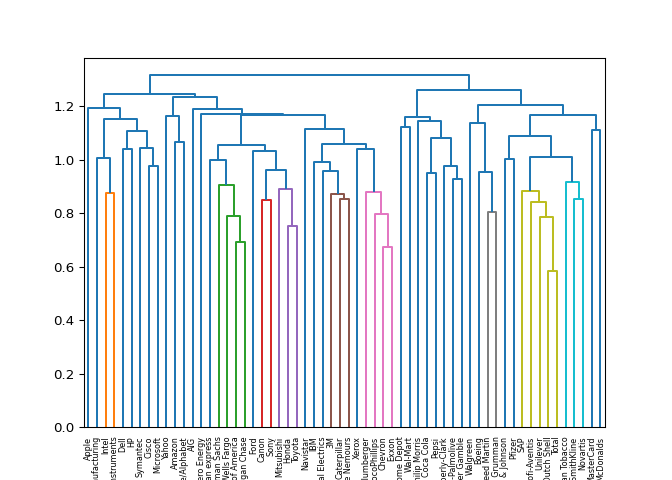
Great work! You can produce great visualizations such as this with hierarchical clustering, but it can be used for more than just visualizations. You’ll find out more about this in the next video!
Cluster labels in hierarchical clustering#
Which clusters are closest?#
In the video, you learned that the linkage method defines how the distance between clusters is measured. In complete linkage, the distance between clusters is the distance between the furthest points of the clusters. In single linkage, the distance between clusters is the distance between the closest points of the clusters.
Consider the three clusters in the diagram. Which of the following statements are true?

A. In single linkage, Cluster 3 is the closest cluster to Cluster 2.
B. In complete linkage, Cluster 1 is the closest cluster to Cluster 2.
Neither A nor B.
A only.
Both A and B.
Different linkage, different hierarchical clustering!#
In the video, you saw a hierarchical clustering of the voting countries
at the Eurovision song contest using ‘complete’ linkage.
Now, perform a hierarchical clustering of the voting countries with
‘single’ linkage, and compare the resulting dendrogram with
the one in the video. Different linkage, different hierarchical
clustering!
You are given an array samples. Each row corresponds to a
voting country, and each column corresponds to a performance that was
voted for. The list country_names gives the name of each
voting country. This dataset was obtained from
Eurovision.
linkage and dendrogram from
scipy.cluster.hierarchy.
samples using the
linkage() function with the method=‘single’
keyword argument. Assign the result to mergings.
country_names as the labels. In addition,
specify the leaf_rotation=90, and
leaf_font_size=6 keyword arguments as you have done
earlier.
# edited/added
eurovision = pd.read_csv("archive/Unsupervised-Learning-in-Python/datasets/eurovision-2016.csv").fillna(0)
scores = pd.crosstab(index=eurovision['From country'], columns=eurovision['To country'], values=eurovision['Televote Points'], aggfunc='first').fillna(12)
samples = scores.values
country_names = list(scores.index)
# Perform the necessary imports
import matplotlib.pyplot as plt
from scipy.cluster.hierarchy import linkage, dendrogram
# Calculate the linkage: mergings
mergings = linkage(samples, method='single')
# Plot the dendrogram
dendrogram(mergings,
labels=country_names,
leaf_rotation=90,
leaf_font_size=6,
)
## {'icoord': [[15.0, 15.0, 25.0, 25.0], [65.0, 65.0, 75.0, 75.0], [85.0, 85.0, 95.0, 95.0], [70.0, 70.0, 90.0, 90.0], [105.0, 105.0, 115.0, 115.0], [80.0, 80.0, 110.0, 110.0], [55.0, 55.0, 95.0, 95.0], [135.0, 135.0, 145.0, 145.0], [185.0, 185.0, 195.0, 195.0], [175.0, 175.0, 190.0, 190.0], [265.0, 265.0, 275.0, 275.0], [255.0, 255.0, 270.0, 270.0], [245.0, 245.0, 262.5, 262.5], [235.0, 235.0, 253.75, 253.75], [315.0, 315.0, 325.0, 325.0], [355.0, 355.0, 365.0, 365.0], [345.0, 345.0, 360.0, 360.0], [335.0, 335.0, 352.5, 352.5], [320.0, 320.0, 343.75, 343.75], [375.0, 375.0, 385.0, 385.0], [405.0, 405.0, 415.0, 415.0], [395.0, 395.0, 410.0, 410.0], [380.0, 380.0, 402.5, 402.5], [331.875, 331.875, 391.25, 391.25], [305.0, 305.0, 361.5625, 361.5625], [295.0, 295.0, 333.28125, 333.28125], [285.0, 285.0, 314.140625, 314.140625], [244.375, 244.375, 299.5703125, 299.5703125], [225.0, 225.0, 271.97265625, 271.97265625], [215.0, 215.0, 248.486328125, 248.486328125], [205.0, 205.0, 231.7431640625, 231.7431640625], [182.5, 182.5, 218.37158203125, 218.37158203125], [165.0, 165.0, 200.435791015625, 200.435791015625], [155.0, 155.0, 182.7178955078125, 182.7178955078125], [140.0, 140.0, 168.85894775390625, 168.85894775390625], [125.0, 125.0, 154.42947387695312, 154.42947387695312], [75.0, 75.0, 139.71473693847656, 139.71473693847656], [45.0, 45.0, 107.35736846923828, 107.35736846923828], [35.0, 35.0, 76.17868423461914, 76.17868423461914], [20.0, 20.0, 55.58934211730957, 55.58934211730957], [5.0, 5.0, 37.794671058654785, 37.794671058654785]], 'dcoord': [[0.0, 9.273618495495704, 9.273618495495704, 0.0], [0.0, 7.211102550927978, 7.211102550927978, 0.0], [0.0, 10.488088481701515, 10.488088481701515, 0.0], [7.211102550927978, 12.0, 12.0, 10.488088481701515], [0.0, 13.114877048604, 13.114877048604, 0.0], [12.0, 13.564659966250536, 13.564659966250536, 13.114877048604], [0.0, 15.874507866387544, 15.874507866387544, 13.564659966250536], [0.0, 14.7648230602334, 14.7648230602334, 0.0], [0.0, 6.782329983125268, 6.782329983125268, 0.0], [0.0, 11.045361017187261, 11.045361017187261, 6.782329983125268], [0.0, 8.94427190999916, 8.94427190999916, 0.0], [0.0, 11.224972160321824, 11.224972160321824, 8.94427190999916], [0.0, 12.083045973594572, 12.083045973594572, 11.224972160321824], [0.0, 12.083045973594572, 12.083045973594572, 12.083045973594572], [0.0, 8.0, 8.0, 0.0], [0.0, 8.366600265340756, 8.366600265340756, 0.0], [0.0, 9.797958971132712, 9.797958971132712, 8.366600265340756], [0.0, 9.899494936611665, 9.899494936611665, 9.797958971132712], [8.0, 10.862780491200215, 10.862780491200215, 9.899494936611665], [0.0, 9.38083151964686, 9.38083151964686, 0.0], [0.0, 10.583005244258363, 10.583005244258363, 0.0], [0.0, 11.224972160321824, 11.224972160321824, 10.583005244258363], [9.38083151964686, 11.224972160321824, 11.224972160321824, 11.224972160321824], [10.862780491200215, 11.313708498984761, 11.313708498984761, 11.224972160321824], [0.0, 11.40175425099138, 11.40175425099138, 11.313708498984761], [0.0, 11.661903789690601, 11.661903789690601, 11.40175425099138], [0.0, 13.416407864998739, 13.416407864998739, 11.661903789690601], [12.083045973594572, 13.711309200802088, 13.711309200802088, 13.416407864998739], [0.0, 14.071247279470288, 14.071247279470288, 13.711309200802088], [0.0, 14.142135623730951, 14.142135623730951, 14.071247279470288], [0.0, 14.142135623730951, 14.142135623730951, 14.142135623730951], [11.045361017187261, 14.491376746189438, 14.491376746189438, 14.142135623730951], [0.0, 14.628738838327793, 14.628738838327793, 14.491376746189438], [0.0, 15.937377450509228, 15.937377450509228, 14.628738838327793], [14.7648230602334, 16.55294535724685, 16.55294535724685, 15.937377450509228], [0.0, 16.911534525287763, 16.911534525287763, 16.55294535724685], [15.874507866387544, 17.204650534085253, 17.204650534085253, 16.911534525287763], [0.0, 17.663521732655695, 17.663521732655695, 17.204650534085253], [0.0, 17.72004514666935, 17.72004514666935, 17.663521732655695], [9.273618495495704, 18.384776310850235, 18.384776310850235, 17.72004514666935], [0.0, 19.79898987322333, 19.79898987322333, 18.384776310850235]], 'ivl': ['Australia', 'Belgium', 'The Netherlands', 'Spain', 'Italy', 'Switzerland', 'Croatia', 'Slovenia', 'Bosnia & Herzegovina', 'Montenegro', 'F.Y.R. Macedonia', 'Serbia', 'Malta', 'France', 'Israel', 'Albania', 'Azerbaijan', 'Bulgaria', 'Cyprus', 'Greece', 'Czech Republic', 'Armenia', 'Germany', 'Russia', 'Moldova', 'Georgia', 'Belarus', 'Ukraine', 'Austria', 'Hungary', 'United Kingdom', 'Ireland', 'Norway', 'Estonia', 'San Marino', 'Latvia', 'Lithuania', 'Denmark', 'Iceland', 'Sweden', 'Finland', 'Poland'], 'leaves': [2, 6, 39, 36, 24, 38, 9, 35, 7, 29, 14, 34, 27, 16, 23, 0, 4, 8, 10, 19, 11, 1, 18, 32, 28, 17, 5, 40, 3, 20, 41, 22, 30, 13, 33, 25, 26, 12, 21, 37, 15, 31], 'color_list': ['C1', 'C2', 'C2', 'C2', 'C2', 'C2', 'C0', 'C0', 'C3', 'C3', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C4', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0', 'C0']}
plt.show()
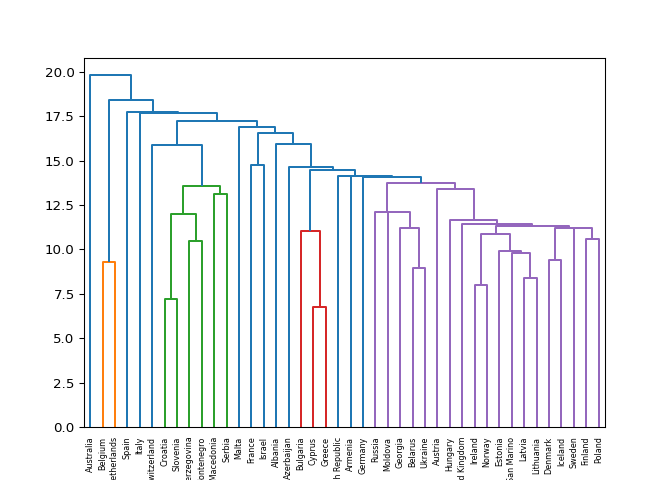
Great work! As you can see, performing single linkage hierarchical clustering produces a different dendrogram!
Intermediate clusterings#
Displayed on the right is the dendrogram for the hierarchical clustering of the grain samples that you computed earlier. If the hierarchical clustering were stopped at height 6 on the dendrogram, how many clusters would there be?

1.
3.
As many as there were at the beginning.
Exactly - great work!
Extracting the cluster labels#
In the previous exercise, you saw that the intermediate clustering of
the grain samples at height 6 has 3 clusters. Now, use the
fcluster() function to extract the cluster labels for this
intermediate clustering, and compare the labels with the grain varieties
using a cross-tabulation.
The hierarchical clustering has already been performed and
mergings is the result of the linkage()
function. The list varieties gives the variety of each
grain sample.
pandas as pd.
fcluster from scipy.cluster.hierarchy.
fcluster() function on mergings. Specify a
maximum height of 6 and the keyword argument
criterion=‘distance’.
df with two columns named
‘labels’ and ‘varieties’, using
labels and varieties, respectively, for the
column values. This has been done for you.
ct between
df\[‘labels’\] and df\[‘varieties’\] to count
the number of times each grain variety coincides with each cluster
label.
# edited/added
samples = np.array(grains.sample(42))[:,:7]
varieties = list(np.array(grains.sample(42))[:,8])
mergings = linkage(samples, method='complete')
# Perform the necessary imports
import pandas as pd
from scipy.cluster.hierarchy import fcluster
# Use fcluster to extract labels: labels
labels = fcluster(mergings, 6, criterion='distance')
# Create a DataFrame with labels and varieties as columns: df
df = pd.DataFrame({'labels': labels, 'varieties': varieties})
# Create crosstab: ct
ct = pd.crosstab(df['labels'], df['varieties'])
# Display ct
print(ct)
## varieties Canadian wheat Kama wheat Rosa wheat
## labels
## 1 1 4 2
## 2 4 6 2
## 3 6 9 8
Fantastic - you’ve now mastered the fundamentals of k-Means and agglomerative hierarchical clustering. Next, you’ll learn about t-SNE, which is a powerful tool for visualizing high dimensional data.
t-SNE for 2-dimensional maps#
t-SNE visualization of grain dataset#
In the video, you saw t-SNE applied to the iris dataset. In this
exercise, you’ll apply t-SNE to the grain samples data and inspect the
resulting t-SNE features using a scatter plot. You are given an array
samples of grain samples and a list
variety_numbers giving the variety number of each grain
sample.
TSNE from sklearn.manifold.
model with
learning_rate=200.
.fit_transform() method of model to
samples. Assign the result to tsne_features.
0 of tsne_features. Assign
the result to xs.
1 of tsne_features. Assign
the result to ys.
xs and
ys. To color the points by the grain variety, specify the
additional keyword argument c=variety_numbers.
# edited/added
variety_numbers = list(np.array(grains.sample(42))[:,7])
# Import TSNE
from sklearn.manifold import TSNE
# Create a TSNE instance: model
model = TSNE(learning_rate=200)
# Apply fit_transform to samples: tsne_features
tsne_features = model.fit_transform(samples)
# Select the 0th feature: xs
xs = tsne_features[:,0]
# Select the 1st feature: ys
ys = tsne_features[:,1]
# Scatter plot, coloring by variety_numbers
plt.scatter(xs, ys, c=variety_numbers)
plt.show()
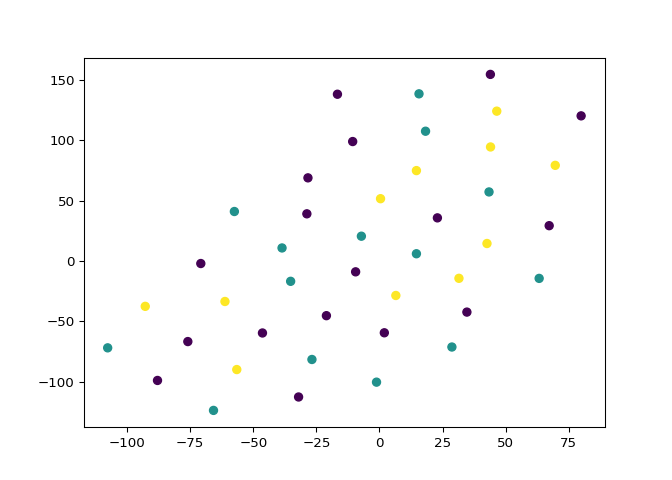
Excellent! As you can see, the t-SNE visualization manages to separate the 3 varieties of grain samples. But how will it perform on the stock data? You’ll find out in the next exercise!
A t-SNE map of the stock market#
t-SNE provides great visualizations when the individual samples can be
labeled. In this exercise, you’ll apply t-SNE to the company stock price
data. A scatter plot of the resulting t-SNE features, labeled by the
company names, gives you a map of the stock market! The stock price
movements for each company are available as the array
normalized_movements (these have already been normalized
for you). The list companies gives the name of each
company. PyPlot (plt) has been imported for you.
TSNE from sklearn.manifold.
model with
learning_rate=50.
.fit_transform() method of model to
normalized_movements. Assign the result to
tsne_features.
0 and column 1 of
tsne_features.
xs and
ys. Specify the additional keyword argument
alpha=0.5.
plt.annotate(), so just hit submit to see the
visualization!
# edited/added
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import normalize
stock = np.array(pd.read_csv("archive/Unsupervised-Learning-in-Python/datasets/company-stock-movements-2010-2015-incl.csv", header = None, skiprows=1))
movements = stock[:,1:]
companies = list(stock[:,0])
normalized_movements = normalize(movements)
# Import TSNE
from sklearn.manifold import TSNE
# Create a TSNE instance: model
model = TSNE(learning_rate=50)
# Apply fit_transform to normalized_movements: tsne_features
tsne_features = model.fit_transform(normalized_movements)
# Select the 0th feature: xs
xs = tsne_features[:,0]
# Select the 1th feature: ys
ys = tsne_features[:,1]
# Scatter plot
plt.scatter(xs, ys, alpha=0.5)
# Annotate the points
for x, y, company in zip(xs, ys, companies):
plt.annotate(company, (x, y), fontsize=5, alpha=0.75)
plt.show()
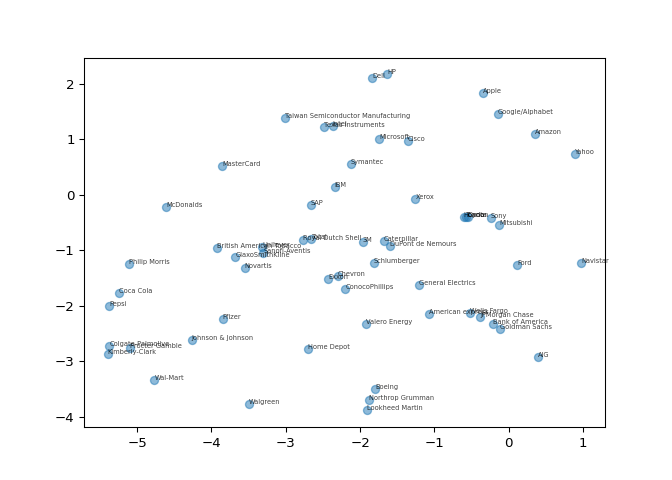
Fantastic! It’s visualizations such as this that make t-SNE such a powerful tool for extracting quick insights from high dimensional data.
